The research paper that cites us is in press in the Journal of Biological Chemistry (reference below). While we were interested in finding as many IGFBP genes in as many vertebrates as we could, studying the characteristics of their genetic code and establishing their evolution, this study is more concerned with the biochemical structures of the actual proteins.
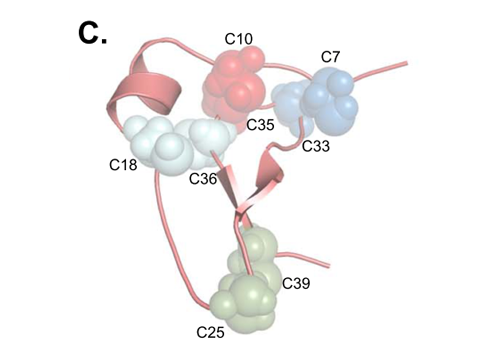
Molecular model of mouse IGFBP-5 (N-terminal), showing the locations of the cysteines that form disulfide bridges. Ref: M. Nili et. al. (see reference below).
Through mass spectrometry, the authors of this study, Mahta Nili and co-workers, have determined the presence of so called disulfide bridges in the IGFBP-5 protein from mouse. Disulfide bridges are formed by the side-chain of the amino acid cysteine and are important structural components of proteins, holding different parts of the three-dimensional structure together and in a way giving the protein part of its "shape". And when it comes to proteins, if you want to understand functions, you have to understand "shape". They could conclude that the structure of the mouse IGFBP-5, looking at the disulfide bridges, probably is very similar to the structure of most other IGFBPs (IGFBP -1,-2,-3 and -4), but not all (IGFBP-6). This is based on comparisons with already known models of a few other IGFBPs as well as our analyses of the amino acid sequences encoded by many different IGFBP genes. Their concluding citation of our paper reads:
Overall, as depicted in Fig. 5C [see above - Daniel], it is likely that the three-dimensional structure of the N-terminal domain of IGFBP-5 is very similar to IGFBP-4, and we predict that IGFBPs 1 - 3 will exhibit analogous structural features.
I agree with their prediction! It's "my" paper they're citing. Our evolutionary analyses would also allow them to say that the structure (probably) holds across all vertebrates, and that it is the ancestral one, but they haven't gone as far as stating that outright.
These are two very different ways of looking at the structure/function relationship of proteins. But at the center of both approaches is trying to connect the biochemical structures of proteins to their functions and their evolution. Each angle feeds the other. Considering all the similarities between IGFBPs I think it's a bit of an evolutionary mystery why we have so many of them - we found some fishes probably have up to 11, us mammals have 6. The structure that the authors of this new study have determined is of the part of IGFBPs (IGFBP-5 in this case) that binds to the Insulin-like Growth Factors (IGFs). This interaction regulates many molecular processed behind our metabolism, cell proliferation and growth, especially during embryonic development, so clearly this functional element is important. But maybe more clues to the evolution of several and diverse IGFBPs lie in other functional elements that have nothing to do with IGFs? Several such biological functions have been described for some IGFBPs, and in our paper from earlier this year we could start suggesting that some of the amino-acids that could underlie these IGF-independent functions show up in some IGFBPs and species, and not in others.
Still, it's only by determining the three-dimensional biochemical structures of proteins that we can start developing an idea about the evolution of functional elements in protein sequences, and how they relate to function.
Nili, M., Mukherjee, A., Shinde, U., David, L., & Rotwein, P. (2011). Defining the disulfide bonds of insulin-like growth factor binding protein-5 by tandem mass spectrometry with electron transfer dissociation and collision induced dissociation Journal of Biological Chemistry DOI: 10.1074/jbc.M111.285528
Ocampo Daza D, Sundström G, Bergqvist CA, Duan C, & Larhammar D (2011). Evolution of the Insulin-Like Growth Factor Binding Protein (IGFBP) Family. Endocrinology, 152 (6), 2278-89 PMID: 21505050
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.