A small announcement: I have an article out as a first author in this month's issue of the journal Endocrinology. It's a nice journal and we spent a long time working on the manuscript so I'm very pleased that it's out. Here's a Worlde word cloud of the whole article... pretty interesting. It sums everything up pretty well actually. It's all about the evolution of the Insulin-like Growth Factor Binding Protein family of genes, or IGFBPs.

Click to see larger.
The IGFBP family genes sit in genomic regions that have long been used to give evidence for the fact that the genome doubled twice early in vertebrate evolution. This in effect makes all extant vertebrates ancient tetraploids; for every one region in the ancient vertebrate genome, there should be 4 corresponding regions in the extant vertebrate genomes. This has become known as the 2R theory. These same genomic regions more famously include the developmentally important HOX gene clusters, which were among the very first indicators that vertebrate genomes are actually ancient tetraploids. See for instance this rewiew or this article.
But even though the IGFBP genes sit in these regions, the studies of their evolution have gone back and forth between different explanations for quite a long time and there is currently a little bit of dispute in the field. At times the IGFBP family has even been used as evidence against the two ancient genome duplications. So what we set out to do when facing this project was to solve the evolutionary history of this gene family once and for all and see if we could say something about the evolution of the different IGFBPs functions. Our analyses can be summarized in this image:
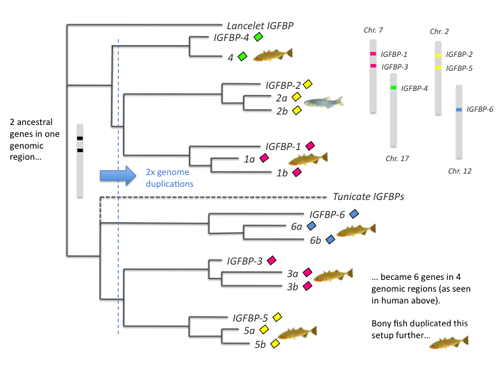
IGFBP evolution through genome duplications. Click to see larger. The lancelet and tunicate are close relatives to vertebrates. The tunicate branch is dotted due to weak support.
The evolutionary relationships between the different IGFBP types fits very well with the 2R theory. A pair of genes in an ancient chromosome region duplicated twice, along with the entire genome, and after some gene losses we get the current setup. This is supported by the evolutionary relationships of several other gene families in the same region.
In the process of finding out the evolutionary history, one also happens to discover new genes quite often. That's always fun! It's especially common for bony fish genomes since the whole group has remnants of yet another ancient genome duplication called 3R. We can also see the results of this third genome duplication in our analyses.
But along with there being a very interesting historical aspect to the analysis of IGFBP evolution when it comes to the genome duplications, there are also functional implications. IGFBPs form part of the growth-promoting hormonal axis by binding to the insulin-like growth factors, or IGFs, whose release into the blood is triggered by the actions of growth hormone. IGFs are also released by most cells in the body directly to the intercellular space acting as local promotors of cell growth and proliferation. So IGFBPs in general modulate the growth-promoting actions of IGFs, but it appears that they do it in very complex ways. Mammals have 6 different IGFBPs, and bony fish have several more. What do they all do? Why do we vertebrates seemingly need so many? Can we say anything about the evolution of their functions simply by looking at the evolution of the genes and proteins? Yes, a little bit. But I leave that for a future blog entry... or you could read the paper.
For now it's enough to say that it's a bit remarkable that throughout the evolutionary history of vertebrates, we have kept so many of the IGFBP genes that have been generated. The fact that the evolutionary history of all these duplicates goes back to the very beginnings of vertebrate evolution say something very interesting about how important their functions are.
Ocampo Daza D, Sundström G, Bergqvist CA, Duan C, & Larhammar D (2011). Evolution of the Insulin-Like Growth Factor Binding Protein (IGFBP) Family. Endocrinology, 152 (6), 2278-89 PMID: 21505050
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.