I've been pottering about in the genome since yesterday morning, looking for the gene families I'm researching in my own work, but the database is still quite rudimentary and tricky to use. Most of the sequences I've searched for come back in fragments, and since the genome hasn't been mapped to chromosomes in the database, it's difficult to find out where in the genome individual sequences are, and to "get to know the neighborhood" of the sequence you're interested in, which is essential for comparative genomics. Thankfully it will (probably) get a more user-friendly interface soon, when it becomes integrated with the Ensembl genome browser, where the other five sequenced fish genomes are already available.
I also made this illustration for my collection of genome species.

The Atlantic cod, Gadus morhua
The basic genome stats reveal a pretty standard vertebrate genome, if there is such a thing. The total (haploid) size is estimated at approx. 830 million base pairs, a bit lower than previous estimates, and the number of identified genes is 22,154 (20,095 protein coding). The closest related fish species with a sequenced genome is the three-spined stickleback, Gasterosteus aculeatus, with a genome of approx. 446 million base pairs and 20,787 identified genes. The best studied fish genome, that of the zebrafish Danio rerio is quite a bit longer, with about 1.5 billion base pairs, but the gene content is similar with about 26,000 identified genes.
Summaries of the main findings from the overall description of the cod genome are available at Nature News and Science Now. These reports highlight the main focus of the published genome description - the loss of genes essential for adaptive immune reactions. The Atlantic cod and several of its closest relatives in the family Gadidae lack genes for the Major Histocompatibility Complex class II (MHC II), one of the proteins that presents antigens to immune cells and initiate an adaptive immune response. They also lack genes for the proteins CD4 and Ii21 (also known as CD74). CD4 is expressed on lymphocytes called helper T-cells and allow them to interact with the MHC II, and Ii21 is involved in the transportation of MHC II proteins to the surface of the cell.
Instead, the Atlantic cod has greatly expanded and diversified its setup of the other class of MHC genes, the MHC class I genes, as well as genes for proteins called Toll-like receptors. These molecules represent another side of immune responses which, seemingly, the cod lineage has recruited to complement its immune defense. It was probably this expansion of MHC I genes and Toll receptor genes that lead to the loss of MHC II, CD4 and Ii21 rather than the other way around.
The researchers confirmed that the genes were missing, by trying to sequence them independently, with no results (which in this case was good). But from a comparative genomics perspective, the evidence they gathered is more compelling. They matched the genomic regions containing these genes in the already known fish genomes against the cod genome sequence and found the corresponding regions.
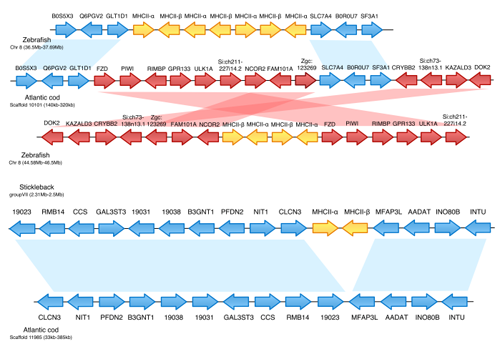
Click on the image to see a larger version. Ref: Supplementary information to B. Star et al. in Nature (see reference below).
As you can see in the image above, the yellow MHC II genes are missing from the corresponding regions of the cod genome, as compared to the zebrafish and stickleback genomes. The same thing can be seen for the Ii21/CD74 genes, but for the CD4 gene they actually found a smaller fragment of the gene in the cod genome, showing that these genes are either going or gone.
These findings helps us understand fish immunity better, and might contribute to improve the aquaculture conditions for cod, as is highlighted in the reports above, but most importantly of all it's a great and interesting example of gene loss and gene duplication, an important mechanism in evolution. In addition, the method I exemplified above is a great example of the type of work you can do once you have several whole genome sequences which you can compare with each other. Once the genomic database is fully functional, many research groups will be able to explore other great questions using similar methods. With this in mind, the cod genome promises to be yet another great contribution in our understanding of how genomes, and therefore organisms, have evolved.
Star, B., Nederbragt, A., Jentoft, S., Grimholt, U., Malmstrøm, M., Gregers, T., Rounge, T., Paulsen, J., Solbakken, M., Sharma, A., Wetten, O., Lanzén, A., Winer, R., Knight, J., Vogel, J., Aken, B., Andersen, Ø., Lagesen, K., Tooming-Klunderud, A., Edvardsen, R., Tina, K., Espelund, M., Nepal, C., Previti, C., Karlsen, B., Moum, T., Skage, M., Berg, P., Gjøen, T., Kuhl, H., Thorsen, J., Malde, K., Reinhardt, R., Du, L., Johansen, S., Searle, S., Lien, S., Nilsen, F., Jonassen, I., Omholt, S., Stenseth, N., & Jakobsen, K. (2011). The genome sequence of Atlantic cod reveals a unique immune system Nature DOI: 10.1038/nature10342
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.